The overarching goal of our research is to understand how genetic variation shapes phenotypic diversity. We study this question in C. elegans and related species. Combining the powerful tools available for studying the C. elegans model with innovative genomic technologies, we strive to elucidate basic principles of how genetic variation shapes our natural world.
Toxin-antidote selfish genes in C. elegans and related species
We like to think that our genes work for us, but sometimes it’s the other way around.
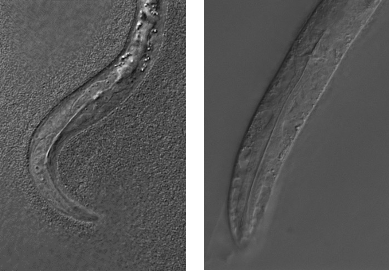
Selfish genes, who do nothing but ensure their survival, are everywhere. Sometimes, we even find them where they’re least expected. During my postdoctoral work, we discovered that a gene long thought to be essential for worm development is, in fact, a selfish gene - part of a toxin-antidote selfish element (Ben-David et al. Science 2017). Toxin-antidote elements cause an individual to kill their offspring, unless they inherit them and express the antidote. For example, the worm shown in the picture (left) was poisoned by a toxin and its pharynx failed to develop. It died shortly after hatching (right picture shows a normal worm hatchling). We have now discovered six more elements in two other worm species (Ben-David et al., Biorxiv 2020). How widespread are these elements in animals? What is their role in evolution? How do they work? These are all open questions we would like to answer.
Key publications
- A maternal-effect selfish genetic element in Caenorhabditis elegans
Science. 356, Issue 6342 (2017)
- Ubiquitous selfish toxin-antidote elements in Caenorhabditis species
bioRxiv (2020)
- Toxin-Antidote Elements Across the Tree of Life
Annual Review of Genetics (2020)
Genetic variation and gene regulation at the cellular level
Genetic variation affects the expression of every gene in our genome.
 We study this relationship at the cellular level
We study this relationship at the cellular level. One of the features that make
C. elegans such a powerful model organism for genetics is that every individual has the same cellular composition. When combined with a cutting-edge genomics tool - single-cell RNA sequencing - we can profile gene expression in the entire worm at cellular resolution. For example, the image on the right shows the clustering of about 50,000
C. elegans cells (Ben-David et al.,
Biorxiv 2020). Different groups (clusters) correspond to different cell types in the worm: muscles, intestinal cells, neurons – it’s all there. We are using this approach to study how genetic variation affects gene expression at the level of single cells. Our ultimate goal is understanding the role of regulatory variation in shaping phenotypes within and between species.
Key publication
- Whole-organism mapping of the genetics of gene expression at cellular resolution
Biorxiv (2020)

