Plasmodium falciparum is the deadliest
form of human malaria, causing the death of over million people annually. The
virulence of P. falciparum is attributed to its ability to evade the
human immune system, by modifying the host red blood cell surface to adhere to
the vascular endothelium and to undergo antigenic variation.
This is achieved by tight regulation of gene
expression that ensures that only a single gene (var) out of a large
repertoire is expressed at a time. Understanding the molecular mechanisms by
which the parasite evades human immune attack could lead to the development of
new drugs that disrupt this ability and would give the human immune system an
opportunity to clear the infection and overcome the disease. The ongoing
research in my lab focuses on the molecular and cellular mechanisms that control
gene expression in P. falciparum.
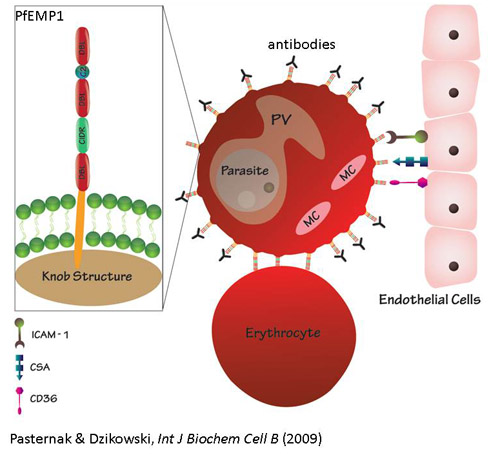
Mechanisms of virulence
gene expression: We are interested to characterize cis and
trans acting elements involved in the epigenetic regulation of var
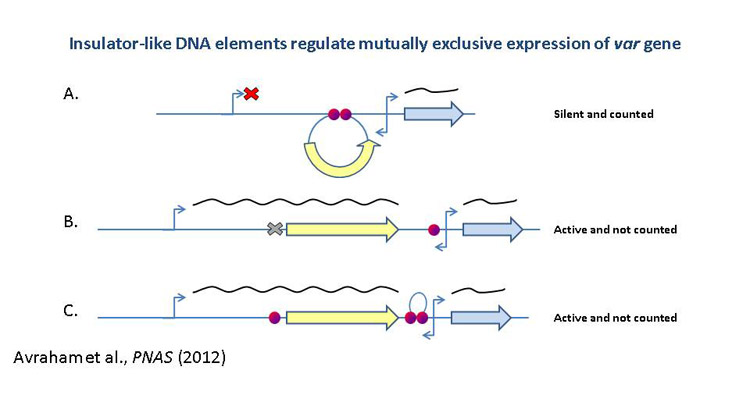
genes. Particularly, identification of the protein/s that specifically
binds the insulator-like DNA elements required for var silencing and
mutually exclusive expression. In addition we found that intronic antisense
ncRNAs are associated with var gene activation and we investigate their
possible role in regulation of these genes. We also study the unknown function
and regulation of the unusual type3 vars. To ease the use of reverse
genetic approaches in P. falciparum we develop new tools to manipulate
gene expression in Plasmodium that could be also used as a novel approach for
drug design.
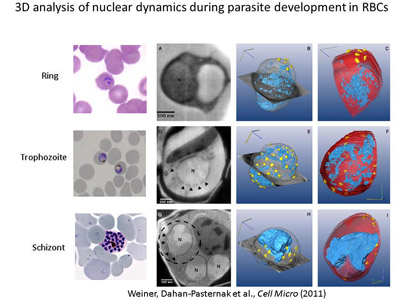
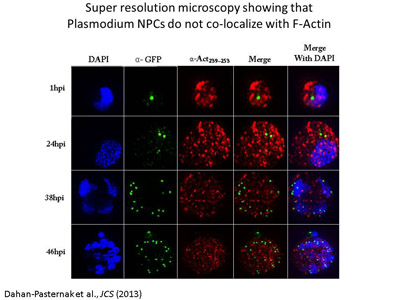
Determination of the role of components
of the nuclear envelope in gene regulation of malaria parasites. In
addition to our efforts that will include whole genome approaches to determine
the possible role of PfSec13 in the nucleoplasm, we identified additional
nucleoporin homologues and we would like to investigate their association with
gene expression in P. falciparum. We currently use bioinformatic,
genetic and biochemical approaches to identify and characterize additional
components of the NE and unveil their role in the parasites' biology.
The role of alternative splicing in
regulating gene expression. Our identification of PfSR1 as the first
alternative splicing factors in the Plasmodium encourage us to take whole genome
approaches to explore its role in regulating gene expression. We created
transgenic parasite that allow induced over expression of PfSR1 and will use
them for Microarray and RNAseq to look at the global changes in transcription
and AS that will enable to understand the importance of such event in parasites'
biology. We currently characterize other candidate SR proteins that were
identifies in our recent paper. In addition, target genes of PfSR1 will be used
to establish artificial mini gene systems that will facilitate exploring
mechanisms of AS in P. falciparum.

