
I grew up in Ramat-Gan and studied Computer Science and Physics, first as an undergraduate and then as a graduate student, at Bar-Ilan University. My research as a PhD student was on stochastic processes and models of information flow in networks. During my PhD, I became interested in biology and worked on biophysical modes. After completing basic training in biology, I joined the labs of Prof. Shula Michaeli at Bar-Ilan University, where I worked on RNA processing in Trypanosomes, and then Prof. Erez. Levanon (RNA editing), where I also completed a short post-doc.
After careful deliberation, I decided to focus on population genetics for my post-doc abroad. Population geneticists study demographic and evolutionary models for genetic variation within individuals in a species. These models are important in medicine, for the purpose of understanding the genetic basis of diseases and traits. In 2011 then, I moved to New York to begin post-doctoral training in the lab of Prof. Itsik Pe’er at Columbia University.
Genetic variation in humans contains important information on biological mechanisms (for example, mutation and recombination rates), historical events and genealogies (when populations mixed or split, or the time of common ancestors), and disease risk factors. My research develops and applies methods from statistics and computer science to extract information encoded in human DNA.
During my stay abroad, I participated in a study of Ashkenazi Jews. Thanks to the recent “DNA sequencing revolution”, it is now possible to efficiently and affordably “read” the DNA sequence of hundreds or even thousands of individuals. In a large-scale effort, we created a catalog of genetic variation from about a hundred Ashkenazi Jews. Our data is now widely used by clinical geneticists, as well as in genetic screening and genetic studies in Israel and abroad. Additionally, our data has demonstrated that Ashkenazi individuals descend from a very small number of founders living only about 700 years ago. We also showed that the Ashkenazi genetic ancestry is part European and part Middle-Eastern, and we mapped the sources and times of gene flow from Europeans into Ashkenazi Jews. (A
link to the article on the genetic catalog).
I returned to Israel in 2015 and joined the faculty of the school of public health at the Hebrew University. In my group, we study population genetics and genetic epidemiology. We develop statistical methods and models to reconstruct historical demographic events in the Israeli and other populations, for example, by using DNA extracted from ancient bones. We also work on estimating the time of origin of Ashkenazi disease mutations. In medicine, we work on expanding the Ashkenazi genetic catalog and on additional applications, such as improving the accuracy of genetic testing before and during pregnancy. In other projects, jointly with university and Hadassah researchers, we study the genetic basis of various diseases and traits.
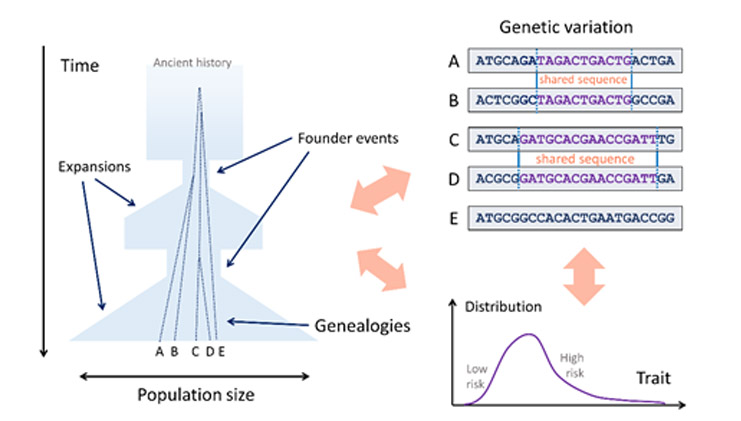
Research in Dr. Carmi’s lab involves demographic events, genealogies, genetic variation, diseases, and traits. Historical changes in the population size (such as due to founder events) affect the time when individuals find their common ancestors. The resulting genealogies affect the extent of genetic variation (for example, the length of shared sequences), and, in turn, the extent of phenotypic differences. The goal of the research is to use genetic variation to improve our understanding of historical demographic events, the genetic basis of traits, and their interplay.

