Research
Vision
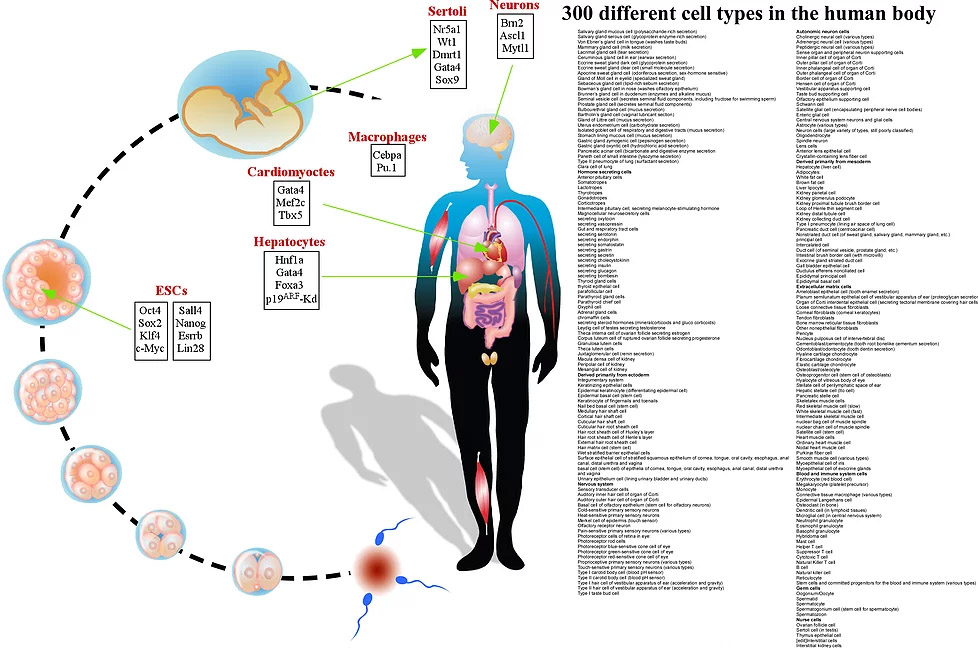
Embryonic stem cells (ESCs) are pluripotent cells derived from the inner cell mass of a blastocyst or earlier morula stage embryos. They are capable of long-term growth; self-renewal and can give rise to every cell, tissue and organ in the fetus's body. When face the right cues ESCs can differentiate to more than 300 different cell types that are the derivatives of the three primary germ layers: ectoderm, endoderm and mesoderm. This complicated and fascinating “skill” raises many intriguing questions. How cell fate decisions are made? What determines cell identity? How external and internal cues facilitate a complete transcriptional and epigenetic resetting? Can we recapitulate ESC plasticity in different somatic cells without reaching the pluripotency state?
To tackle these questions and to illuminate the fundamental elements that regulate cell plasticity, we are exploiting cutting-edge single-cell technologies, genome engineering methods, mouse genetic manipulations and sophisticated bioinformatic approaches.

Read more...
Regenerative medicine is a new and expanding area that aims to replace lost or damaged cells, tissues or organs in the human body through cellular transplantation. Embryonic stem cells (ESCs) are pluripotent cells that are capable of long-term growth; self-renewal and can give rise to every cell, tissue and organ in the fetus's body. The “hyperdynamic” chromatin state, which characterizes the ESC epigenetic state, facilitates a rapid and efficient reaction to external and internal cues that lead to the activation of key master regulators that drive the cells into their developmental fate (1-5). Thus, ESCs hold great promise for cell therapy as a source of diverse differentiated cell types. Two major bottlenecks to realizing such potential are allogenic immune rejection of ESC-derived cells by recipients and ethical issues. In 2006, two Japanese scientists, Takahashi and Yamanaka, changed the way we used to think about cell plasticity when they showed that introduction of four transcription factors, Oct4, Sox2, Klf4 and Myc (OSKM), can reprogram adult fibroblasts into functional embryonic stem-like cells (also termed induced pluripotent stem cells (iPSCs)) (6,7). Resetting the epigenome of a somatic cell to a pluripotent state has already been achieved by somatic cell nuclear transfer (SCNT), but this was ensued by approximately 8000 genes that are expressed within the oocyte (8,9). The notion that as little as four factors are sufficient to reset the epigenome of a cell, opened a new avenue where scientists have attempted to convert different adult cells into other somatic cell types from ontogenetically different lineages, by avoiding the pluripotent state, using a specific subset of key master regulators (10,11). Several subsets of cell types such as hematopoietic cells, different neuronal cells cardiomyocytes, hepatocytes, embryonic Sertoli cells, endothelial cells and RPE were converted from different somatic cells by employing the direct conversion approach. Importantly, the generation of iPSCs and directly converted cells resolve the problems of allogenic immune rejection of ESC-derived cells by recipients and ethical issues. However, the majority of the directly converted cells are not stable and represent mostly incomplete reprogramming state and the vast majority of the resulting iPSCs, although could activate their endogenous circuitry, exhibit poor developmental potential in mice. This suggests that the current prevailing reprogramming methods are not ideal and must be improved before considering applying the converted cells in the clinic.
To improve the quality of the converted cells, the molecular mechanisms that underlie the various conversion processes must be uncovered. Two major obstacles in deciphering the elements responsible for the various reprogramming processes are the heterogeneity of the transduced cell population and the small fraction of cells that eventually will be converted. These obstacles yield two major challenges: (1) The ability to detect and capture early on the rare cells that eventually will be converted and (2) to monitor the transcriptional profile of cells at the single-cell level.
To overcome these limitations, our group focuses on several conversion models and by employing sophisticated engineered fluorescent reporter systems and innovative single-cell technologies we show that major elements that dictate successful reprogramming events and aid in improving the quality of the converted cells, can be illuminated.
Uncovering these elements and improving the quality of the converted cells will bring the regenerative medicine field one-step closer toward the clinic.
Models
Induced Pluripotent Stem Cells (iPSCs)
Since the discovery that introduction of four transcription factors, OSKM, into somatic cells can initiate a reprogramming process that eventually yields iPSCs (7), there has been a remarkable progress in characterizing the resulting iPSCs. It is clear today that the quality of the iPSCs is one of the major obstacles that must be resolved before applying iPSCs to the clinic. In contrast to ESCs, the quality of iPSCs varies widely between different colonies, and a large proportion of these colonies is of low pluripotency potential, as measured by their poor capability to contribute to chimeras and to generate a healthy “all iPSC" mouse using tetraploid complementation assay. Over the past seven years, a significant effort has been made to improve reprogramming efficiency, and to examine the transcriptome, proteome, epigenome, and the pluripotency potential of iPSCs. These studies revealed a large number of genetic and epigenetic aberrations throughout the genome of iPSCs, that are distinct from those found in ESCs (1). Despite those efforts, we still lack a clear understanding of how the mechanisms to create iPSCs influence the quality of iPSCs.
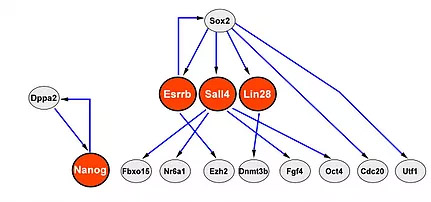
In our previous work, we have analyzed the expression of 48 genes in single cells at various stages during the reprogramming process (12,13). Analysis of early stages revealed considerable variation in gene expression between cells in contrast to late stages. We found that Esrrb, Utf1, Lin28, and Dppa2 have a better prediction potential for cells to become induced pluripotent stem cells (iPSCs) than expression of previously suggested reprogramming markers such as Fbxo15, Fgf4, and Oct4. We noticed that stochastic gene expression early in reprogramming is followed by a late deterministic phase with Sox2 being the upstream factor in the gene expression network. This network allows us to reprogram cells to iPSCs with the absence of “Yamanaka” key pluripotency genes and Nanog, thereby demonstrating for the first time that the core pluripotency circuitry can be activated from different entries.
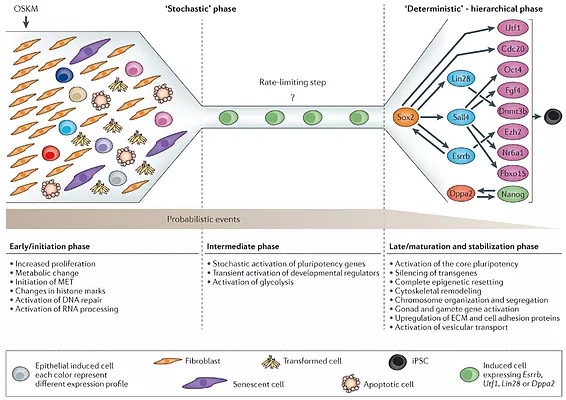
In a follow-up study, we reasoned that a combination of key factors derived from this later phase will reprogram cells in a more controlled way and therefore might uniformly yield iPSCs of high quality. We chose Sall4, Esrrb and Lin28 key downstream players during the late reprogramming phase and Nanog because it participated in a separate pathway. Indeed, we noticed that ectopic expression of Sall4, Nanog, Esrrb and Lin28 (SNEL) in mouse embryonic fibroblasts (MEFs) generates iPSCs of superior quality compared to iPSCs derived from OSKM (14). SNEL-iPSCs contributed to high-grade chimaeras and produced “all-iPSC” mice by 4n complementation at a significantly higher frequency than OSKM-derived iPSCs. These results demonstrate that bioinformatic models derived from in vitro single-cell data can aid in improving the quality of the converted cells.
Induced embryonic Sertoli-like Cells (ieSCs)
Sertoli cells are considered the "supporting cells" of the testis that play an essential role in sex determination during embryogenesis and in spermatogenesis during adulthood (15). Their essential roles in male fertility along with their immunosuppressive and neurotrophic properties make them an attractive cell type for therapeutic applications. Therefore, we decided to convert fibroblasts into Sertoli-like cells. We characterized the role of specific transcription factor combinations in the transition from fibroblasts to induced embryonic Sertoli-like cells (ieSCs) and identify key steps in the process. Initially, transduced fibroblasts underwent a mesenchymal to epithelial transition (MET) and then acquired the ability to aggregate, formed tubular-like structures, and expressed embryonic Sertoli-specific markers (16). These cells integrated into testicular cords in gonad explants and support germ cell and neural progenitor cell survival in vitro, thus making them a desirable cell type for fertility and neurodegenerative treatments. Importantly, these fundamental insights gained during the formation of Sertoli-like cells could not be addressed using conventional in vitro and in vivo approaches because endogenous Sertoli cells cannot be sustained in culture and KO of each of the key master regulators in vivo caused early lethality or the absence of the gonad when using Cre-specific approach.
These studies support the ideology that transcription factor-mediated cell fate conversions mimic physiological cell fate transitions and suggest that somatic cell conversion by defined factors is not only useful for generating cell types that are relevant for therapeutic applications but rather also a very powerful research tool to address basic questions in biology and to resolve issues that are difficult to address using conventional in vivo and in vitro approaches (17).
Induced Trophoblast Stem Cells (iTSCs)
iPSCs undergo extensive nuclear reprogramming and are generally indistinguishable from embryonic stem cells (ESCs) in their functional capacity and transcriptome and DNA methylation profiles. However, the direct conversion of cells from one lineage to another often yields incompletely reprogrammed, functionally compromised cells, raising the question of whether pluripotency is required to achieve a high degree of nuclear reprogramming. Here, we show that transient expression of Gata3, Eomes, and Tfap2c in mouse fibroblasts induces stable, transgene-independent trophoblast stem cells (iTSCs). iTSCs possess transcriptional profiles highly similar to blastocyst-derived TSCs, with comparable methylation and H3K27ac patterns and genome-wide H2A.X deposition (18). iTSCs generate trophoectodermal lineages upon differentiation, form hemorrhagic lesions, and contribute to developing placentas in chimera assays, indicating a high degree of nuclear reprogramming, with no evidence of passage through a transient pluripotent state. Together, these data demonstrate that extensive nuclear reprogramming can be achieved independently of pluripotency (18).
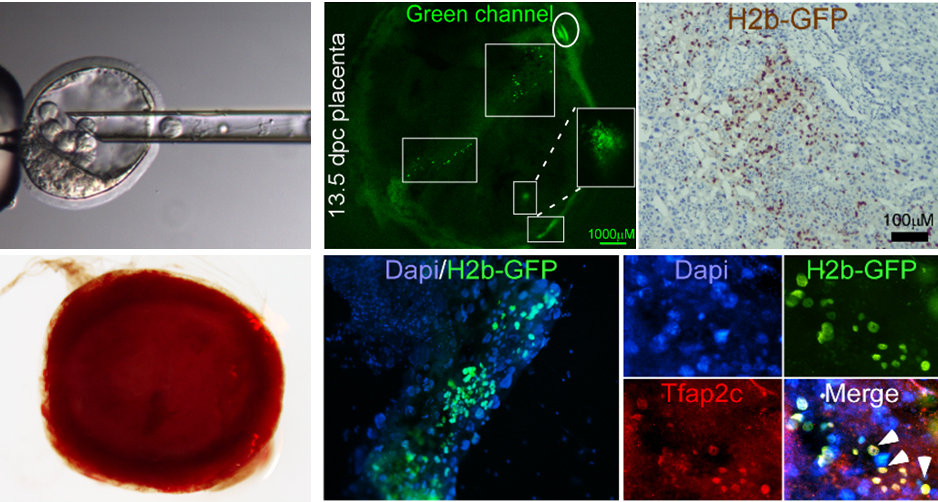
The placenta is the least understood human organ, but arguably one of the most important ones. It influences not only the health of a woman and her fetus during pregnancy, but also their lifelong health. Placental insufficiency occurs when the placenta does not develop properly, or is damaged. Placental dysfunction diseases are associated with low birth weight, premature birth, and birth defects. One such disease is fetal growth restriction (FGR, also termed intrauterine growth restriction [IUGR]), that tends to display mild mental retardation and in severe cases causes fetal death. They also carry increased risk of complications for the mother. To date, tools to model or treat these diseases are limited because all attempts to isolate and propagate the human placenta precursor cells (i.e. trophoblast stem cells, TSCs) in the dish have failed. Here, our main goal is to identify the conditions required to maintain TSCs in culture in their undifferentiated state and to investigate the molecular mechanisms that underlie the fibroblast-to-iTSC conversion process.
References
1. Buganim, Y., Faddah, D. A., and Jaenisch, R. (2013) Mechanisms and models of somatic cell reprogramming. Nat Rev Genet.
2. Meshorer, E., Yellajoshula, D., George, E., Scambler, P. J., Brown, D. T., and Misteli, T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell
3. Soufi, A., Donahue, G., and Zaret, K. S. (2012) Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell
4. Zaret, K. S., and Carroll, J. S. (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev
5. Zhu, J., Adli, M., Zou, J. Y., Verstappen, G., Coyne, M., Zhang, X., Durham, T., Miri, M., Deshpande, V., De Jager, P. L., Bennett, D. A., Houmard, J. A., Muoio, D. M., Onder, T. T., Camahort, R., Cowan, C. A., Meissner, A., Epstein, C. B., Shoresh, N., and Bernstein, B. E. (2013) Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell
6. Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell
7. Takahashi, K., and Yamanaka, S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell
8. Gurdon, J. B., and Melton, D. A. (2008) Nuclear reprogramming in cells. Science
9. Young, R. A. (2011) Control of the embryonic stem cell state. Cell
10. Graf, T., and Enver, T. (2009) Forcing cells to change lineages. Nature
11. Lee, T. I., and Young, R. A. (2013) Transcriptional regulation and its misregulation in disease. Cell
12. Buganim, Y., Faddah, D. A., Cheng, A. W., Itskovich, E., Markoulaki, S., Ganz, K., Klemm, S. L., van Oudenaarden, A., and Jaenisch, R. (2012) Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell
13. Pan, G., and Pei, D. (2012) Order from chaos: single cell reprogramming in two phases. Cell Stem Cell
14. Buganim, Y., Markoulaki, S., Wietmarschen, VN., Hoke, H., Wu, T., Ganz, K., Akhtar-Zaidi, B., He, Y., Abraham, B.J., Porubsky, D., Kulenkampff, E., Faddah, D.A., Shi, L., Gao, Q., Sarkar,S., Cohen, M., Goldmann, J., Nery, J.R., Schultz, M.J., Ecker, J.R., Xiao, A., Young, R.A., Lansdorp, P.M., Jaenisch, R. (2014) The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell
15. Skinner, M. K., and Griswold, M. D. (2005) Sertoli cell biology. Elsevier Academic Press
16. Buganim, Y., Itskovich, E., Hu, Y. C., Cheng, A. W., Ganz, K., Sarkar, S., Fu, D., Welstead, G. G., Page, D. C., and Jaenisch, R. (2012) Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell
17. Buganim, Y., and Jaenisch, R. (2012) Transdifferentiation by defined factors as a powerful research tool to address basic biological questions. Cell Cycle
18. Benchetrit H, Herman S, van Wietmarschen N, Wu T, Makedonski K, Maoz N, Yom Tov N, Stave D, Lasry R, Zayat V, Xiao A, Lansdorp PM, Sebban S, Buganim Y. (2015) Extensive Nuclear Reprogramming Underlies Lineage Conversion into Functional Trophoblast Stem-like Cells. Cell Stem Cell

