Virulence, Organelle Biology and Molecular Genetics of Malaria Parasites
Malaria is a leading cause of morbidity and mortality worldwide, infecting nearly 250 million people and killing about 450,000 people every year. It is caused by obligate eukaryotic parasites of the genus Plasmodium but it is one species, P. falciparum, that is responsible for most of the mortality. As yet, there are no effective vaccines against malaria and the parasite has gained resistance to all clinically available antimalarial drugs. Therefore, it is vital that we constantly generate new drugs and identify potential drug targets to stay ahead of this deadly disease.
Developing advanced genetics tools to the study of a non-model organism
Despite its clinical significance, P. falciparum remains a difficult-to-study non-model organism with a limited set of molecular tools, a challenge restricting our understanding of fundamental parasitic processes. The impact of our research group stems from our ability and motivation to develop and apply advanced molecular-genetic and biochemical techniques for the study of unique parasitic phenomena. These include CRISPR/Cas9-mediated gene editing, various conditional-knockdown systems, marker-less genetic modifications, new expression systems and other novel technologies. With these advanced tools in hand, we ask questions that address fascinating biological problems in original ways.

The ancient Apicoplast of P. falciparum
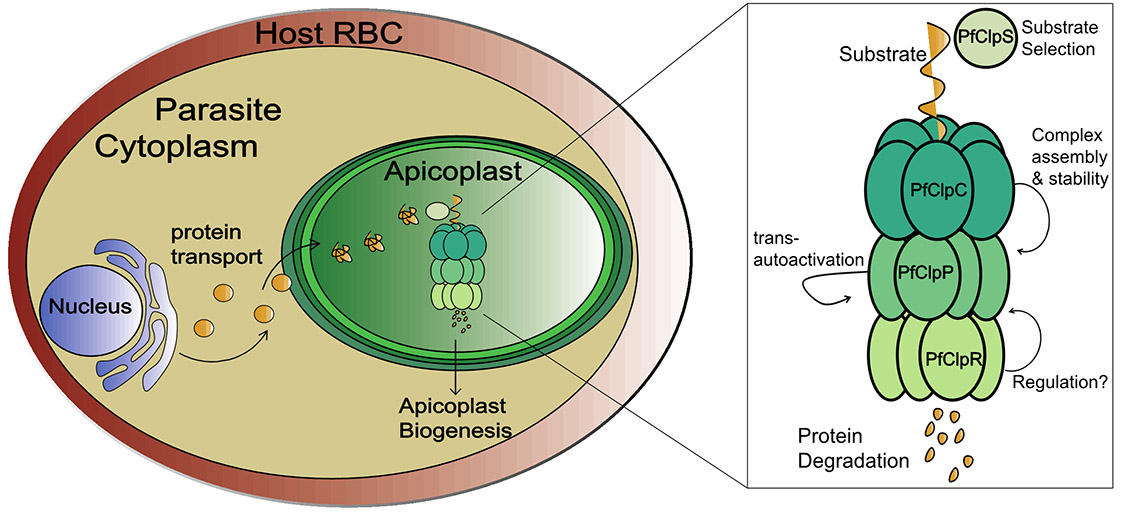
P. falciparum contains a unique organelle known as the apicoplast, a distinctive endosymbiont with its own genetic material. The apicoplast harbors essential, parasite-specific metabolic pathways, and drugs targeting the apicoplast have shown success in the clinic. Most of these drugs target the organelle's prokaryotic-derived translation system, but, in fact, less than 10% of the apicoplast proteome is encoded and synthesized by the organelle itself. Most of the apicoplast proteins are encoded and regulated by the cell nucleus and it is unclear how the apicoplast controls its own proteome and biogenesis. In our research we discovered that a Clp (caseinolytic proteases) proteolytic system that resides within the apicoplast functions as a key regulator of organelle biology. We hypothesize that the plastid is using Clp-mediated degradation as the main mechanism to control protein levels and the derived organellar functions.
The Clp system in malaria Parasites
In bacteria, the evolutionary ancestors of the apicoplast, Clp proteins play vital roles in cell division, protein homeostasis and virulence. Consequentially, they have been in the center of antibacterial drug discovery programs. Typically, they form a proteolytic complex where a ClpP protease is paired with a Clp chaperone. Several putative Clp proteins have been localized to the apicoplast of P. falciparum but their function is completely unknown. Using a plethora of cellular assays including microscopy, qRT-PCR and imaging-flow cytometry, we have shown that the chaperone PfClpC is essential for parasite viability and apicoplast function. Novel techniques including regulatable mutants, dominant-negative and new expression systems, revealed molecular interactions between components of the Clp system, and its role as a master regulator of plastid proteome. Our current and future goals are:
- Study Clp regulation and proteolytic capacity in the apicoplast
- Identify Clp substrates that are involved in organelle biogenesis
- Repurpose bacterial Clp inhibitors as anti-malarials